import pandas as pd
import os
import csv
import io
import numpy as npsegment_id = 78
filename = f'temperature_degree_c/{segment_id}_temperature_degree_c.csv'
df = pd.read_csv(filename, parse_dates=['timestamp_utc'], index_col='timestamp_utc', date_format="%m/%d/%Y %I:%M:%S %p")
# todo: figure out why the data is always sorted
df = df.sort_index()Make missing gap¶
percentage = 10
num_rows = len(df)
num_missing = int(num_rows * percentage / 100)
# Ensure at least one row is set as missing
num_missing = max(1, num_missing)
# Randomly select a starting index for the contiguous block
start_index = np.random.randint(0, num_rows - num_missing + 1)
df['missing'] = df.iloc[:,0]
# Set the contiguous block of rows as missing (NaN)
df.iloc[start_index:start_index + num_missing, df.columns.get_loc('missing')] = np.nan
df_original = df.copy()df.plot()<Axes: xlabel='timestamp_utc'>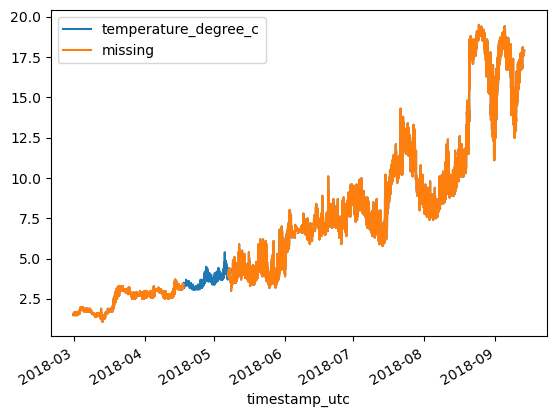
Last Observation Carried Forward¶
df = df_original.copy()
df['missing'] = df['missing'].ffill()
df.plot()<Axes: xlabel='timestamp_utc'>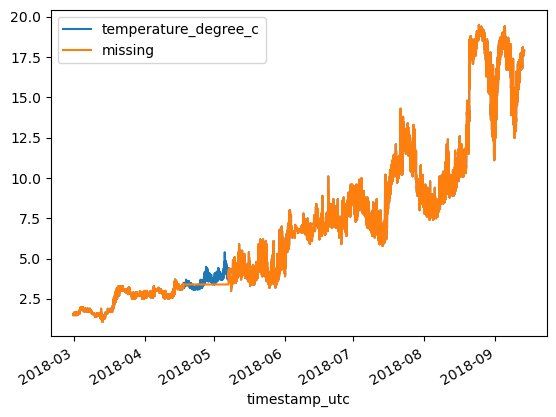
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.0349 RMSE: 0.145
Mean Value¶
df = df_original.copy()
df['missing'] = df['missing'].fillna(df['missing'].mean())
df.plot()<Axes: xlabel='timestamp_utc'>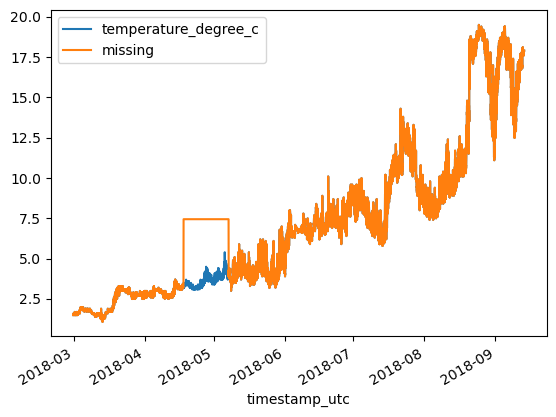
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.38 RMSE: 1.21
Linear Interpolation¶
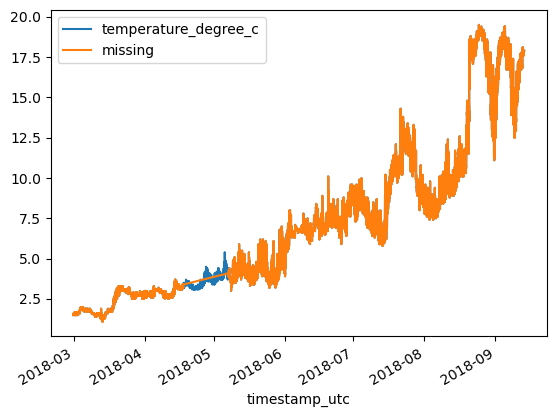
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='linear')
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.0238 RMSE: 0.0919
Nearest Neighbour¶
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='nearest')
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.0234 RMSE: 0.093
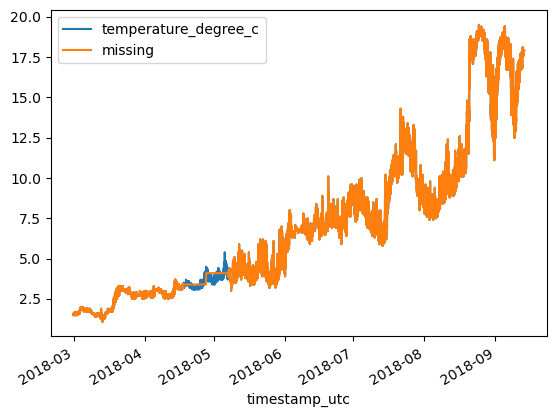
Polynomial Interpolation¶
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='polynomial', order=2)
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 3.31 RMSE: 12.8
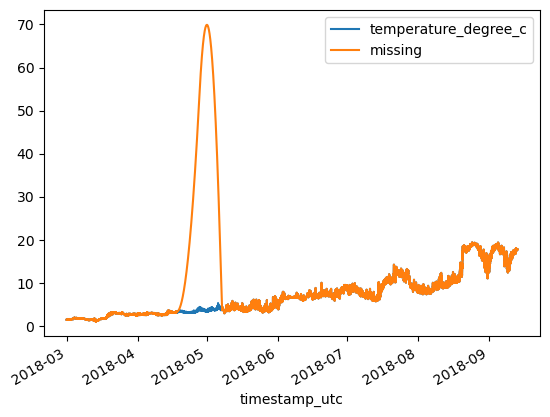
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='polynomial', order=3)
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 4.04 RMSE: 14.9
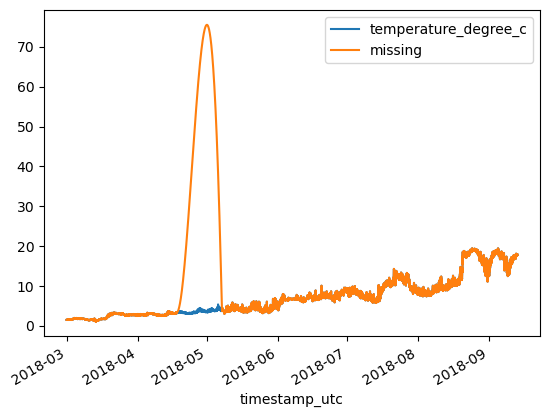
Spline interpolation¶
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='spline', order=2)
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.032 RMSE: 0.13
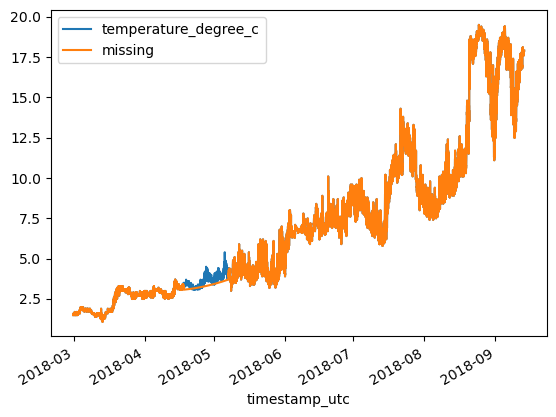
df = df_original.copy()
df['missing'] = df['missing'].interpolate(method='spline', order=3)
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.0292 RMSE: 0.12
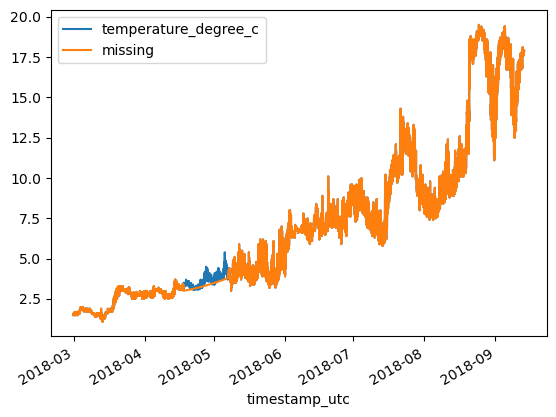
Moving average imputation¶
df = df_original.copy()
df['missing'] = df['missing'].rolling(4).mean()
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')MAE: 0.0971 RMSE: 0.165
MissForest¶
df = df_original.copy()
from missforest import MissForest
# Initialize the magical forest
imputer = MissForest()
# Impute away
df_imputed = imputer.fit_transform(df)
df_imputed.plot()
df.plot()
df['error'] = df['temperature_degree_c'] - df['missing']
MAE = np.mean(abs(df['error']))
RMSE = np.sqrt(np.mean((df['error'])**2))
print(f'MAE: {MAE:.3}\tRMSE: {RMSE:.3}')/home/jmunroe/miniforge3/lib/python3.12/site-packages/missforest/missforest.py:333: UserWarning: Label encoding is no longer performed by default. Users will have to perform categorical features encoding by themselves.
warnings.warn("Label encoding is no longer performed by default. "
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[17], line 9
6 imputer = MissForest()
8 # Impute away
----> 9 df_imputed = imputer.fit_transform(df)
11 df_imputed.plot()
13 df.plot()
File ~/miniforge3/lib/python3.12/site-packages/missforest/missforest.py:596, in MissForest.fit_transform(self, x)
583 def fit_transform(self, x: pd.DataFrame = None) -> pd.DataFrame:
584 """Calls class methods `fit` and `transform` on `x`.
585
586 Parameters
(...)
594 Imputed dataset (features only).
595 """
--> 596 return self.fit(x).transform(x)
File ~/miniforge3/lib/python3.12/site-packages/missforest/missforest.py:364, in MissForest.fit(self, x)
362 _validate_infinite(x.drop(self._categorical, axis=1))
363 else:
--> 364 _validate_infinite(x)
366 self._numerical = [c for c in x.columns if c not in self._categorical]
368 # Sort column order according to the amount of missing values
369 # starting with the lowest amount.
File ~/miniforge3/lib/python3.12/site-packages/missforest/_validate.py:115, in _validate_infinite(x)
102 def _validate_infinite(x: pd.DataFrame):
103 """Checks if argument `x` contains infinite values.
104
105 Parameters
(...)
113 - If argument `x` contains infinite values.
114 """
--> 115 if np.any(np.isinf(x)):
116 raise ValueError("Argument `x` must not contains infinite values.")
File ~/miniforge3/lib/python3.12/site-packages/pandas/core/generic.py:2171, in NDFrame.__array_ufunc__(self, ufunc, method, *inputs, **kwargs)
2167 @final
2168 def __array_ufunc__(
2169 self, ufunc: np.ufunc, method: str, *inputs: Any, **kwargs: Any
2170 ):
-> 2171 return arraylike.array_ufunc(self, ufunc, method, *inputs, **kwargs)
File ~/miniforge3/lib/python3.12/site-packages/pandas/core/arraylike.py:407, in array_ufunc(self, ufunc, method, *inputs, **kwargs)
402 if method == "__call__" and not kwargs:
403 # for np.<ufunc>(..) calls
404 # kwargs cannot necessarily be handled block-by-block, so only
405 # take this path if there are no kwargs
406 mgr = inputs[0]._mgr
--> 407 result = mgr.apply(getattr(ufunc, method))
408 else:
409 # otherwise specific ufunc methods (eg np.<ufunc>.accumulate(..))
410 # Those can have an axis keyword and thus can't be called block-by-block
411 result = default_array_ufunc(inputs[0], ufunc, method, *inputs, **kwargs)
File ~/miniforge3/lib/python3.12/site-packages/pandas/core/internals/managers.py:361, in BaseBlockManager.apply(self, f, align_keys, **kwargs)
358 kwargs[k] = obj[b.mgr_locs.indexer]
360 if callable(f):
--> 361 applied = b.apply(f, **kwargs)
362 else:
363 applied = getattr(b, f)(**kwargs)
File ~/miniforge3/lib/python3.12/site-packages/pandas/core/internals/blocks.py:393, in Block.apply(self, func, **kwargs)
387 @final
388 def apply(self, func, **kwargs) -> list[Block]:
389 """
390 apply the function to my values; return a block if we are not
391 one
392 """
--> 393 result = func(self.values, **kwargs)
395 result = maybe_coerce_values(result)
396 return self._split_op_result(result)
TypeError: ufunc 'isinf' not supported for the input types, and the inputs could not be safely coerced to any supported types according to the casting rule ''safe''Kalman smoothing imputation¶
from rpy2.robjects.packages import importr
imputeTS = importr("imputeTS")
from rpy2 import robjects
kalman_StructTs = robjects.r['na.kalman']
kalman_auto_arima = robjects.r['na.kalman']df = df_original.copy()
#kalman_StructTs(df['missing'], model='StructTS')KNN¶
from fancyimpute import knndf = df_original.copy()
#knn.KNN(k=1).fit_transform(df[['missing']])