import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import holoviews as hv
hv.extension('bokeh')df = pd.read_csv('dataset.csv', parse_dates=True, index_col=0)Here is a dataset with missing values.
df.sample(10)We can see that the data set has several regions of missing data.
plt.imshow(df.T, aspect='auto', interpolation='nearest')
plt.xlabel('Time')
plt.ylabel('Site')
plt.show()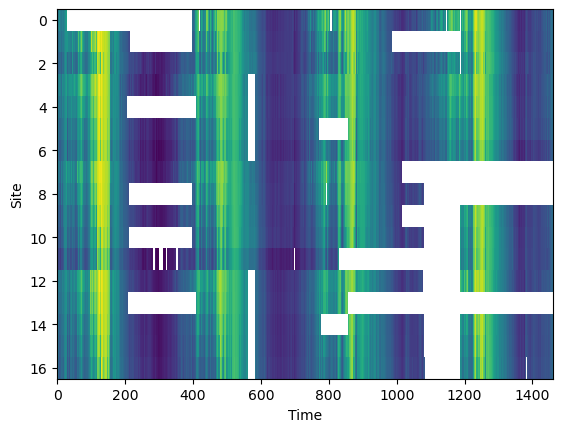
We wish to examine different imputation algorithms to see how they compare with each other. In this study we consider KNN, MICE, MissForest, and a simple linear univariate method. The process is adaptable to other imputation based methods.
def missing_data_fraction(df):
X = df.values
N = X.size
m = np.isnan(X).sum()
missing_data = m / N * 100
return missing_dataprint (f'Original missing data {missing_data_fraction(df):0.1f}%')Original missing data 17.9%
We take our dataset at restrict it the study sites where 90% of the data is available. Once we have validated that the imputation algorithm are working as expected we will come back and use the best one to fill in more of the missing data in the dataset.
# Calculate the percentage of non-missing data for each study site
non_missing_percentage = df.notna().mean() * 100
# Filter study sites with at least 90% non-missing data
selected_sites = non_missing_percentage[non_missing_percentage >= 90].index
df_selected = df[selected_sites]plt.imshow(df_selected.T, aspect='auto', interpolation='nearest')
plt.xlabel('Time')
plt.ylabel('Site')
plt.show()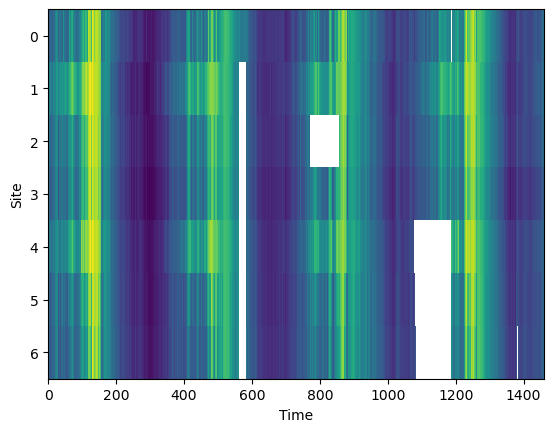
print (f'Missing data {missing_data_fraction(df_selected):0.1f}% for "ground truth"')Missing data 5.3% for "ground truth"
We will refer to this dataset as the true dataset for comparison to the estimated reconstructions. Notice that even this dataset has some missing data.
df_true = df_selected.copy()Introduce artifical gaps¶
Prepare the dataset by introducing artificial gaps. We can randomly select a fixed number of sites and introduce gaps of a fixed length.
def add_artificial_gaps(df, num_sites=1, gap_length=30):
np.random.seed(4152)
gaps = {}
# randomly set a n-day contiguous segment as missing for each column
random_columns = np.random.choice(df.columns, size=num_sites, replace=False)
N = len(df.values.flatten())
m = df.isnull().values.flatten().sum()
missing_data = m / N * 100
for col in random_columns:
# Randomly select the start of the n-day segment
start_idx = np.random.randint(0, len(df) - gap_length)
end_idx = start_idx + gap_length
gaps[col] = [start_idx, end_idx]
# Set the values in this range to NaN
df.iloc[start_idx:end_idx, df.columns.get_loc(col)] = np.nan
df = df_true.copy()
add_artificial_gaps(df, 7, 90)plt.imshow(df.T, aspect='auto', interpolation='nearest')
plt.xlabel('Time')
plt.ylabel('Site')
plt.show()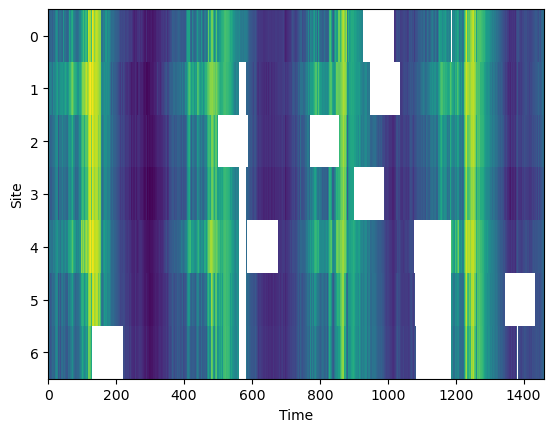
print (f'Artificial missing data {missing_data_fraction(df):0.1f}%')Artificial missing data 11.3%
We can vary the parameters of the number of sites and gap length to explore how the different imputation algorithms perform with different about of missing data.
Imputation methods¶
Simple Imputation (mean)¶
Scikit-Learn has a general approach to using imputation algorithms that we will mirror for all subsequent imputation algorithms.
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(missing_values=np.nan, strategy='mean')
# Fit and transform the dataset
df_imputed = pd.DataFrame(imputer.fit_transform(df), columns=df.columns, index=df.index)This imputer replaces missing data with the mean of all data in the record.
df_imputeddictionary = { i: hv.Overlay( [
hv.Curve(df_true[str(i)], label='Truth'),
hv.Curve(df_imputed[str(i)], label='Imputed'),
hv.Curve(df[str(i)], label='Artificial Gap'),
]) for i in df_true.columns}
hv.HoloMap(dictionary, kdims='Study Site').opts(aspect=2, title='Simple Imputer (mean)')The visualization above allow inpsection of how the imputation algorithm is working at each study site. The ‘true’ value is shown in blue and the imputed value is shown in red. Every site has a randomly chosen 90-day interval removed somewhere in the time series.
Any regions with red lines but no blue lines indicate missing gaps in the original dataset.
To evaluate this fit we compare back with the truth. We can calculate the Mean Absolute Error and the Root Mean Square Error.
error = df_imputed - df_true
MAE = np.mean(abs(error))
RMSE = np.sqrt(np.mean((error)**2))
print(f'MAE = {MAE:.2f}, RMSE = {RMSE:.2f}')MAE = 0.20, RMSE = 0.94
We will compare different methods in terms of these metrics and as function of the relative fraction of missing data.
Linear interpolation¶
This is a univariate method where we infill the missing data with a straight line between boundary points on any interval.
df_imputed = df.interpolate(method='linear')dictionary = { i: hv.Overlay( [
hv.Curve(df_true[str(i)], label='Truth'),
hv.Curve(df_imputed[str(i)], label='Imputed'),
hv.Curve(df[str(i)], label='Artificial Gap'),
]) for i in df_true.columns}
hv.HoloMap(dictionary, kdims='Study Site').opts(aspect=2, title='Linear Interpolation')error = df_imputed - df_true
MAE = np.mean(abs(error))
RMSE = np.sqrt(np.mean((error)**2))
print(f'MAE = {MAE:.2f}, RMSE = {RMSE:.2f}')MAE = 0.07, RMSE = 0.37
The error is reduced considerable compared to impute with the simple imputer using the mean.
KNN¶
For K Nearest Neighbours (KNN), a hyperparameter is the number of neighbour to use. We will tune this hyperparameter in a subsequent notebook.
from sklearn.impute import KNNImputer
imputer = KNNImputer(n_neighbors=7)
df_imputed = pd.DataFrame(imputer.fit_transform(df), columns=df.columns, index=df.index)dictionary = { i: hv.Overlay( [
hv.Curve(df_true[str(i)], label='Truth'),
hv.Curve(df_imputed[str(i)], label='Imputed'),
hv.Curve(df[str(i)], label='Artificial Gap'),
]) for i in df_true.columns}
hv.HoloMap(dictionary, kdims='Study Site').opts(aspect=2, title='KNN')error = df_imputed - df_true
MAE = np.mean(abs(error))
RMSE = np.sqrt(np.mean((error)**2))
print(f'MAE = {MAE:.2f}, RMSE = {RMSE:.2f}')MAE = 0.02, RMSE = 0.11
MICE¶
Multiple Imputation by Chained Equation (MICE)
from statsmodels.imputation.mice import MICEData
mice_data = MICEData(df, k_pmm=3)
n_iterations = 10
mice_data.update_all(n_iterations)
df_imputed = pd.DataFrame(mice_data.data.values, columns=df.columns, index=df.index)dictionary = { i: hv.Overlay( [
hv.Curve(df_true[str(i)], label='Truth'),
hv.Curve(df_imputed[str(i)], label='Imputed'),
hv.Curve(df[str(i)], label='Artificial Gap'),
]) for i in df_true.columns}
hv.HoloMap(dictionary, kdims='Study Site').opts(aspect=2, title='MICE')error = df_imputed - df_true
MAE = np.mean(abs(error))
RMSE = np.sqrt(np.mean((error)**2))
print(f'MAE = {MAE:.2f}, RMSE = {RMSE:.2f}')MAE = 0.02, RMSE = 0.09
MissForest¶
MissForest is an imputation method based on Random Forests. The implementation used here is a reimplementation in Python of the algorithm from the original paper which in turn wraps the random forest regressor from scikit-learn. There are other MissForest implementations available in PyPi but I was not able to get them to run successfully.
from imputeMF import imputeMFMissForest is a non-parametric method but we can set a maximum number of iterations. An optimum value for the number of iteration is explored in a subsequent notebook.
df_imputed = pd.DataFrame(imputeMF(df.values, 5, print_stats=True), columns=df.columns, index=df.index)Statistics:
iteration 1, gamma = 0.014922669733387309
Statistics:
iteration 2, gamma = 0.00023221646561239455
Statistics:
iteration 3, gamma = 1.2199979642092591e-05
Statistics:
iteration 4, gamma = 6.9909467418157684e-06
Statistics:
iteration 5, gamma = 6.595499472639891e-06
dictionary = { i: hv.Overlay( [
hv.Curve(df_true[str(i)], label='Truth'),
hv.Curve(df_imputed[str(i)], label='Imputed'),
hv.Curve(df[str(i)], label='Artificial Gap'),
]) for i in df_true.columns}
hv.HoloMap(dictionary, kdims='Study Site').opts(aspect=2, title='MissForest')error = df_imputed - df_true
MAE = np.mean(abs(error))
RMSE = np.sqrt(np.mean((error)**2))
print(f'MAE = {MAE:.2f}, RMSE = {RMSE:.2f}')MAE = 0.01, RMSE = 0.07