import pandas as pd
import os
import csv
import io
import numpy as np
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import seaborn as sns
from matplotlib.dates import DateFormatter
import sklearn.impute
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from missforest import MissForest
Imputation¶
Next steps include implementing goodness-of-fit (GoF) indicators ( coefficient of determination, percent bias, Kling-Gupta efficiency, MAE, RMSE, ) to evaluate the performance of the imputation algorithm. An experiment should be replicated many times to present the GoF as a distribution. Then explore how that GoF changes with the amount of missing data (gap sizes) and tuning of hyperparameters. These GoF indicators can then also be compared to other imputation algorithms to determine if MissForest (or any other more computational involved algorithm) is actually “better” than simple univariate methods.
The goal for the next two weeks is to set up the framework and run the experiments:
- Measurement of GoF indicators
- Multiple replicates of each experiment
- As a function of continuous gap size
- As a function of number of randomly removed samples
- Compare to ‘simple’ methods such as replace with mean or median or linear interpolation
Goodness-of-fit indicators¶
Coefficient of determination ()¶
variable = 'temperature_degrees'
df = pd.read_csv('data.csv', parse_dates=True, index_col=0)
# Define the cutoff date
cutoff_date = "2020-09-01"
# Select rows past the cutoff date
df = df[df.index >= pd.Timestamp(cutoff_date)]
# Define the cutoff date
cutoff_date = "2024-08-31"
# Select rows past the cutoff date
df = df[df.index <= pd.Timestamp(cutoff_date)]
# Calculate the percentage of non-missing data for each study site
non_missing_percentage = df.notna().mean() * 100
# Filter study sites with at least 50% non-missing data
selected_sites = non_missing_percentage[non_missing_percentage >=99].index
df_selected = df[selected_sites]def plot_data_available(df):
# Creating a boolean DataFrame where True indicates non-missing data
non_missing_data = ~df.isna()
# Plotting
plt.figure(figsize=(12, 8))
sns.heatmap(non_missing_data.T, xticklabels= 180, cmap=['white', 'black'], cbar=False)
plt.xlabel('Time')
plt.ylabel('Study Site ID')
plot_data_available(df_selected)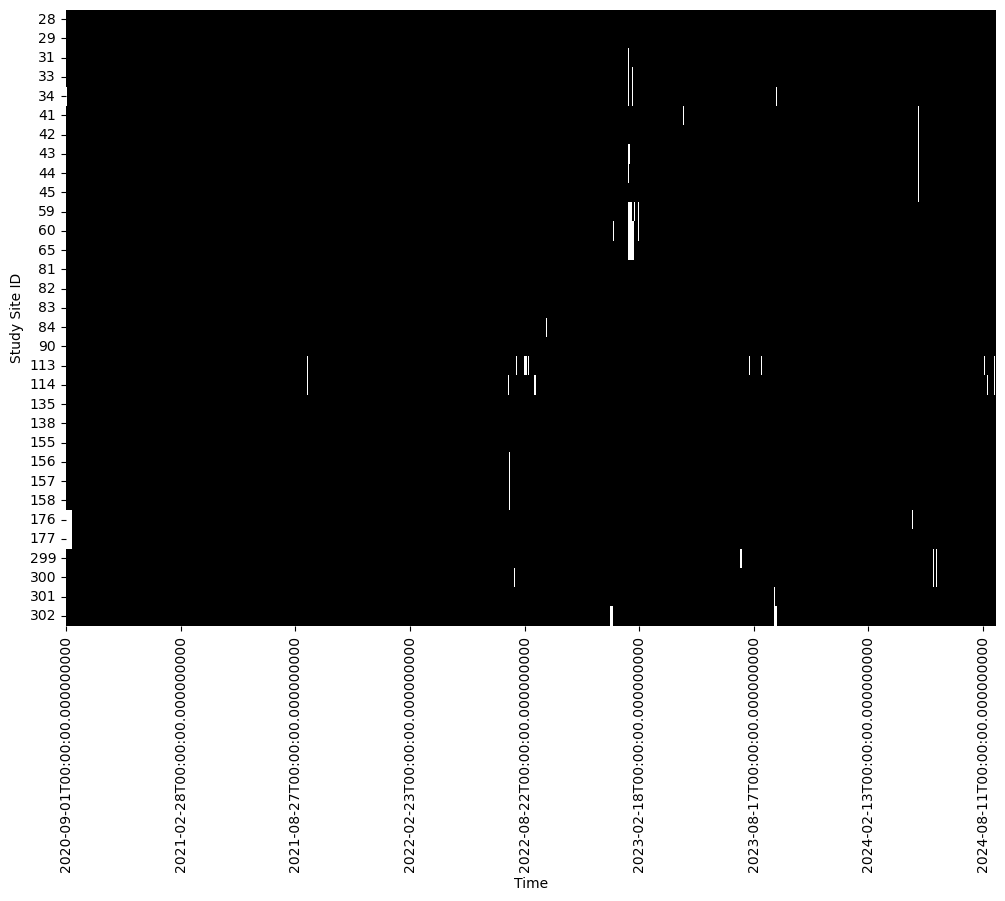
Dataset consists of 31 “stations” with 1461 observations each.
Remove contiguous segments¶
At each station remove contiguous segments (having lengths 7, 14, 21, 30 days)
df = df_selected.copy()
# Function to randomly set a n-day contiguous segment as missing for each column
n = 180
for col in df.columns:
# Randomly select the start of the n-day segment
start_idx = np.random.randint(0, len(df) - n)
end_idx = start_idx + n
# Set the values in this range to NaN
df.iloc[start_idx:end_idx, df.columns.get_loc(col)] = np.nanplot_data_available(df)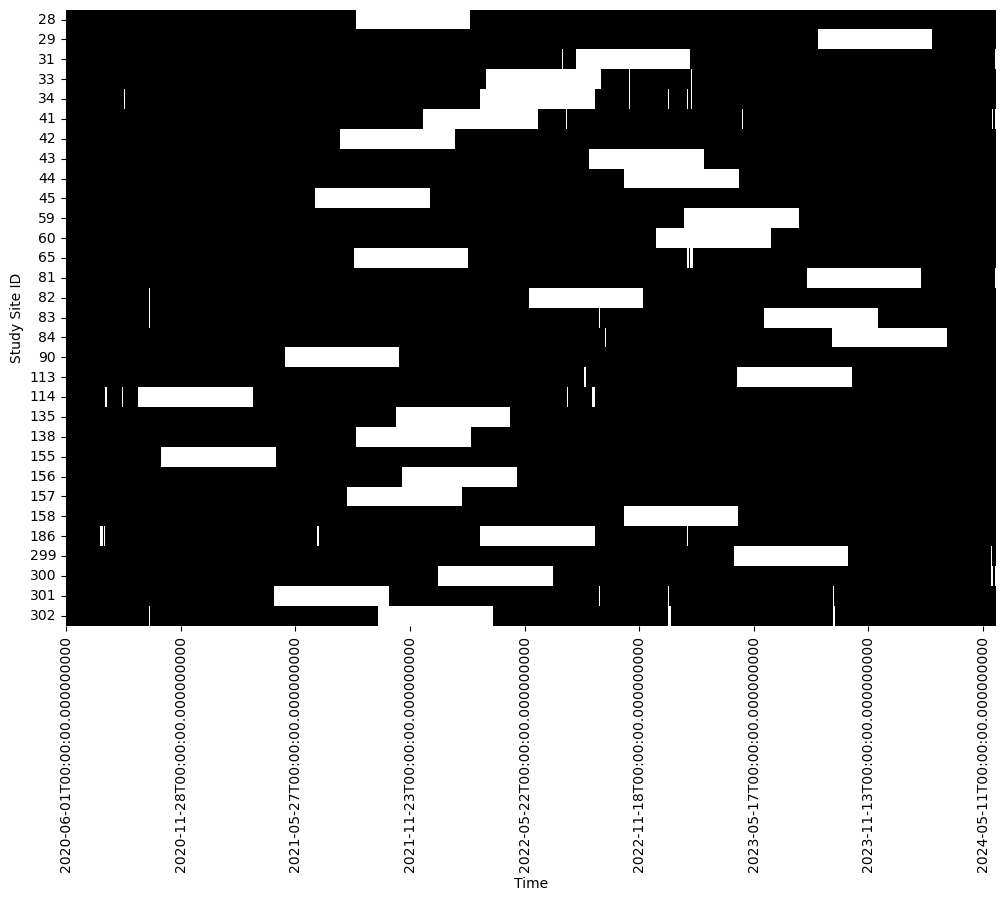
def run_experiment(method = 'MF', gap_type = 'continuous', n = 1):
variable = 'temperature_degrees'
df = pd.read_csv('data.csv', parse_dates=True, index_col=0)
# Define the cutoff date
cutoff_date = "2020-06-01"
# Select rows past the cutoff date
df = df[df.index >= pd.Timestamp(cutoff_date)]
# Define the cutoff date
cutoff_date = "2024-06-01"
df = df[df.index < pd.Timestamp(cutoff_date)]
# Calculate the percentage of non-missing data for each study site
non_missing_percentage = df.notna().mean() * 100
# Filter study sites with at least 50% non-missing data
selected_sites = non_missing_percentage[non_missing_percentage >= 99].index
df_selected = df[selected_sites]
# artifical gaps
df = df_selected.copy()
if gap_type == 'continuous':
# randomly set a n-day contiguous segment as missing for each column
for col in df.columns:
# Randomly select the start of the n-day segment
start_idx = np.random.randint(0, len(df) - n)
end_idx = start_idx + n
# Set the values in this range to NaN
df.iloc[start_idx:end_idx, df.columns.get_loc(col)] = np.nan
elif gap_type == 'random':
# remove randomly selected n days as missing for each column
for col in df.columns:
# Randomly sample unique dates
random_dates = np.random.choice(df.index, size=n, replace=False)
# Set the values to NaN for those dates
df.loc[random_dates, col] = np.nan
else:
raise Exception(f'Unknown gap_type {gap_type}')
if method == 'MF':
# Imputer
clf = RandomForestClassifier(n_jobs=-1)
rgr = RandomForestRegressor(n_jobs=-1, n_estimators=5)
imputer = MissForest(clf, rgr, verbose=1,max_iter=5)
df_sim = imputer.fit_transform(df)
elif method == 'linear':
df_sim = df.interpolate(method='linear')
else:
raise Exception(f'Unknown method {method}')
suffix = f'_{gap_type}_{n}_{method}'
# save output
df_selected.to_csv(f'obs{suffix}.csv')
df_imputed.to_csv(f'sim{suffix}.csv')
method = 'linear'
gap_type = 'continuous'
for n in [7, 14, 21, 30, 60, 180, 365]:
run_experiment(gap_type=gap_type, n=n, method=method)
gap_type = 'random'
for n in [30, 60, 90, 120, 180, 365]:
run_experiment(gap_type=gap_type, n=n, method=method)
station_id = '28'
plt.figure(figsize=(16,4))
plt.plot(df_imputed[station_id], label='Simulated')
plt.plot(df_selected[station_id], label='Observed')
plt.plot(abs(df_selected[station_id] - df_imputed[station_id])+20, label='Error')
plt.legend()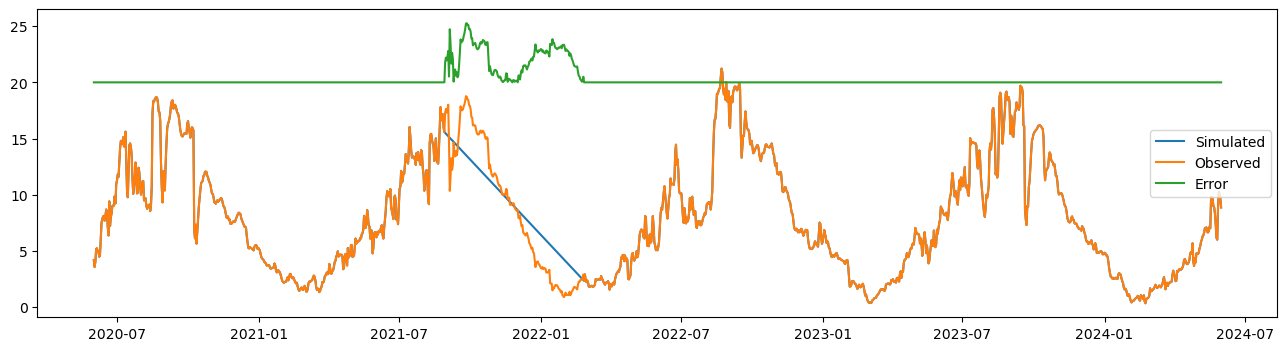
Calculate GoFs¶
S = df_imputed
O = df_selected
μo = O.mean()
σo = O.std()
μs = S.mean()
σs = S.std()
# bias ratio
β = μs / μo
# variability ratio
γ = (σs/μs) / (σo/μo)
## correlation coefficient
r = df_selected.corrwith(df_imputed)R2 = ((O - μo) * (S - μs)).sum() / (np.sqrt(((O-μo)**2).sum()) * np.sqrt(((S-μs)**2).sum()))
PBIAS = (S - O).sum() / O.sum() * 100
KGE = 1 - np.sqrt((r - 1)**2 + (β - 1)**2 + (γ - 1)**2)
def KGE(S, O):
μo = O.mean()
σo = O.std()
μs = S.mean()
σs = S.std()
# bias ratio
β = μs / μo
# variability ratio
γ = (σs/μs) / (σo/μo)
## correlation coefficient
r = O.corrwith(S)
return 1 - np.sqrt((r - 1)**2 + (β - 1)**2 + (γ - 1)**2)
fig, ax = plt.subplots(1, 3, figsize=(12, 4))
ax[0].boxplot(R2)
ax[0].set_title('$R^2$')
ax[1].boxplot(PBIAS)
ax[1].set_title('PBIAS (%)')
ax[2].boxplot(KGE)
ax[2].set_title('KGE')
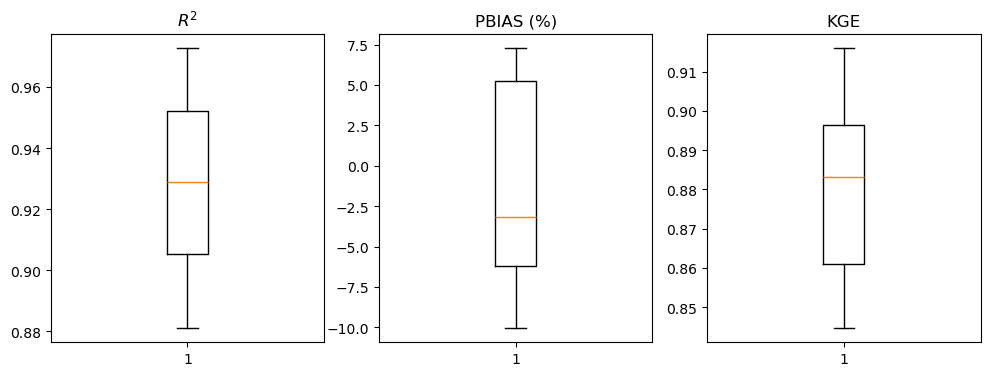
def gof(O, S):
μo = O.mean()
σo = O.std()
μs = S.mean()
σs = S.std()
# bias ratio
β = μs / μo
# variability ratio
γ = (σs/μs) / (σo/μo)
## correlation coefficient
r = O.corrwith(S)
R2 = ((O - μo) * (S - μs)).sum() / (np.sqrt(((O-μo)**2).sum()) * np.sqrt(((S-μs)**2).sum()))
PBIAS = (S - O).sum() / O.sum() * 100
KGE = 1 - np.sqrt((r - 1)**2 + (β - 1)**2 + (γ - 1)**2)
return pd.DataFrame({'R2': R2, 'PBIAS': PBIAS, 'KGE': KGE})performance = {}
gap_type = 'continuous'
performance[gap_type] = { 'R2': {}, 'PBIAS': {}, 'KGE': {} }
for n in [7, 14, 21, 30, 60, 180, 365]:
for method in ['', '_linear']:
suffix = f'_{gap_type}_{n}{method}'
S = pd.read_csv(f'sim{suffix}.csv', index_col=0)
O = pd.read_csv(f'obs{suffix}.csv', index_col=0)
results = gof(S, O)
if method == '':
method = 'MF'
elif method == '_linear':
method = 'LR'
position = f'{n} {method}'
performance[gap_type]['R2'][position] = results['R2']
performance[gap_type]['PBIAS'][position] = results['PBIAS']
performance[gap_type]['KGE'][position] = results['KGE']
gap_type = 'random'
performance[gap_type] = { 'R2': {}, 'PBIAS': {}, 'KGE': {} }
for n in [30, 60, 90, 120, 180, 365]:
for method in ['', '_linear']:
suffix = f'_{gap_type}_{n}{method}'
S = pd.read_csv(f'sim{suffix}.csv', index_col=0)
O = pd.read_csv(f'obs{suffix}.csv', index_col=0)
results = gof(S, O)
if method == '':
method = 'MF'
elif method == '_linear':
method = 'LR'
position = f'{n} {method}'
performance[gap_type]['R2'][position] = results['R2']
performance[gap_type]['PBIAS'][position] = results['PBIAS']
performance[gap_type]['KGE'][position] = results['KGE']
for gap_type in ['continuous', 'random']:
for metric in ['R2', 'PBIAS', 'KGE']:
performance[gap_type][metric] = pd.DataFrame(performance[gap_type][metric])performance['continuous']['R2']fig, ax = plt.subplots(3, 2, figsize=(12, 16))
for i, gap_type in enumerate(['continuous', 'random']):
labels = list(performance[gap_type]['R2'].columns)
ax[0,i].boxplot(performance[gap_type]['R2'])
ax[0,i].set_xticklabels(labels, rotation=90)
ax[0,i].set_title('$R^2$')
ax[1,i].boxplot(performance[gap_type]['PBIAS'])
ax[1,i].set_xticklabels(labels, rotation=90)
ax[1,i].set_title('PBIAS (%)')
ax[2,i].boxplot(performance[gap_type]['KGE'])
ax[2,i].set_xticklabels(labels, rotation=90)
ax[2,i].set_title('KGE')
plt.show()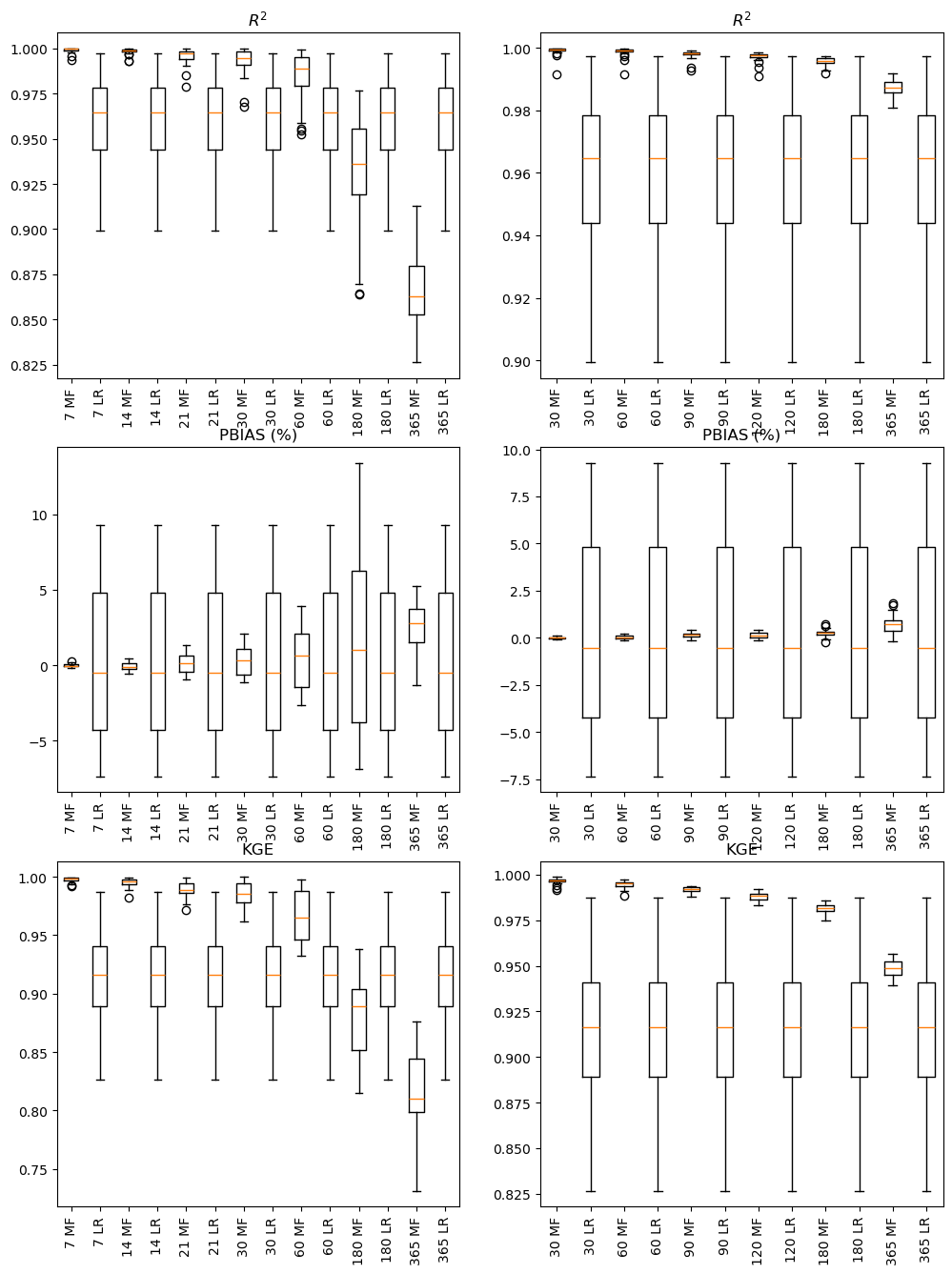
results = gof(S, O)results.boxplot()<Axes: >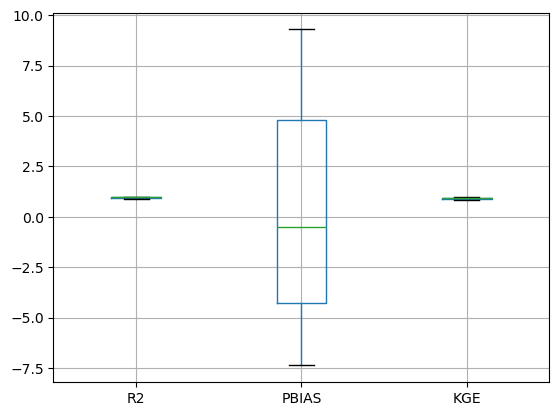
plt.boxplot(gof(S,O), positions)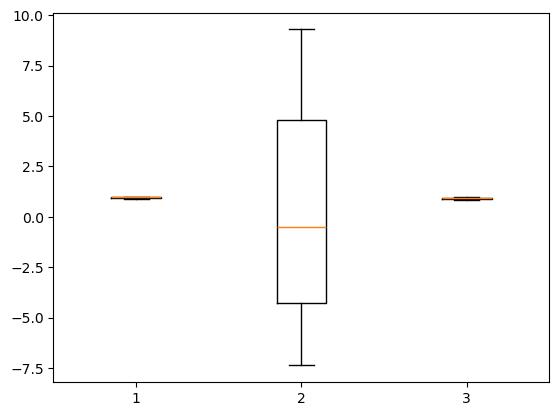
fig, ax = plt.subplots(3, 2, figsize=(12, 16))
ax[0].boxplot(R2)
ax[0].set_title('$R^2$')
ax[1].boxplot(PBIAS)
ax[1].set_title('PBIAS (%)')
ax[2].boxplot(KGE)
ax[2].set_title('KGE')---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
Cell In[270], line 3
1 fig, ax = plt.subplots(3, 2, figsize=(12, 16))
----> 3 ax[0].boxplot(R2)
4 ax[0].set_title('$R^2$')
6 ax[1].boxplot(PBIAS)
AttributeError: 'numpy.ndarray' object has no attribute 'boxplot'