import pandas as pd
import os
import numpy as np
import matplotlib.pyplot as plt
from experiments import run_experiment, KNNImputer, MICEImputer
from rich.progress import trackThis notebook is used to explore how different hyperparameters work for different imputation algorithms.
Determine hyperparameter n for KNN¶
We need to select the number of nearest neighbours to use for KNN.
num_sites = 3
gap_length = 90
dataset = 'dataset.csv'
ns = [2,3,5,7,9,11,13,15,17,19,21]
results_list = []
filtered_results = []
for n in track(ns):
for i in range(30):
result = run_experiment(KNNImputer(n_neighbors=n),
minimum_missing_data=90,
num_sites=num_sites,
gap_length=gap_length,
rerun_experiment=True,
dataset=dataset, random_seed=None)
results_list.append(result)
filtered_result = {k: v for k, v in result.items() if isinstance(v, (int, float, str))}
filtered_result['n'] = n
filtered_results.append(filtered_result)
filtered_results = pd.DataFrame(filtered_results)Loading...
averaged_results = filtered_results[['n', 'RMSE', 'MAE']].groupby('n').agg({
'MAE': ['mean', 'std'], 'RMSE': ['mean', 'std']}).reset_index()
rmse_mean = averaged_results[('RMSE', 'mean')]
rmse_std = averaged_results[('RMSE', 'std')]
mae_mean = averaged_results[('MAE', 'mean')]
mae_std = averaged_results[('MAE', 'std')]fig, ax = plt.subplots(2, 1, figsize=(8, 6))
ax[0].errorbar(ns, mae_mean, yerr=mae_std, fmt='o', capsize=4, label='MAE')
ax[0].set_title('MAE vs n with Error Bars')
ax[0].set_xlabel('n')
ax[0].set_ylabel('MAE')
ax[0].legend()
ax[1].errorbar(ns, mae_mean, yerr=mae_std, fmt='o', capsize=4, label='MAE')
ax[1].set_xlabel('n')
ax[1].set_ylabel('RMSE')
ax[1].legend()
ax[0].set_title(f'Error vs $n$ for KNN')
ax[0].set_xticks([])
ax[1].set_xticks(ns)
plt.show()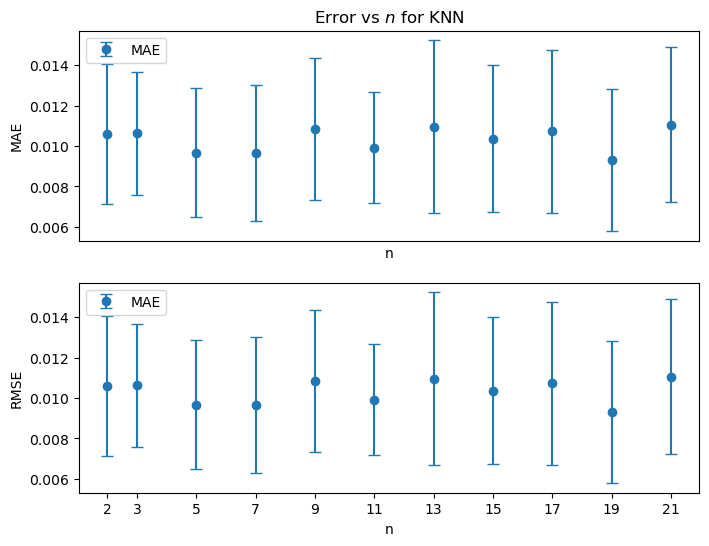
There is no clear trend to indicate a “best” value for when using KNN for the the Shelburne County Dataset (for the experiment of randomly introducing artificial 90-day gaps into three of the time series in the set.)
In analyses of other datasets as long as then KNN appears to give similar results. We choose as the default for the number of neighbors in KNN.
Determine hyperparameter k_pmm for MICE¶
num_sites = 3
gap_length = 90
dataset = 'dataset.csv'
k_pmms = [2,3,5,7,9,11]
results_list = []
filtered_results = []
for k_pmm in track(k_pmms):
for i in range(3):
result = run_experiment(MICEImputer(k_pmm),
minimum_missing_data=90,
num_sites=num_sites,
gap_length=gap_length,
rerun_experiment=True,
dataset=dataset, random_seed=None)
results_list.append(result)
filtered_result = {k: v for k, v in result.items() if isinstance(v, (int, float, str))}
filtered_result['k_pmm'] = k_pmm
filtered_results.append(filtered_result)
filtered_results = pd.DataFrame(filtered_results)Loading...
averaged_results = filtered_results[['k_pmm', 'RMSE', 'MAE']].groupby('k_pmm').agg({
'MAE': ['mean', 'std'], 'RMSE': ['mean', 'std']}).reset_index()
rmse_mean = averaged_results[('RMSE', 'mean')]
rmse_std = averaged_results[('RMSE', 'std')]
mae_mean = averaged_results[('MAE', 'mean')]
mae_std = averaged_results[('MAE', 'std')]fig, ax = plt.subplots(2, 1, figsize=(8, 6))
ax[0].errorbar(k_pmms, mae_mean, yerr=mae_std, fmt='o', capsize=4, label='MAE')
ax[0].set_xlabel('k_pmms')
ax[0].set_ylabel('MAE')
ax[0].legend()
ax[1].errorbar(k_pmms, mae_mean, yerr=mae_std, fmt='o', capsize=4, label='MAE')
ax[1].set_xlabel('k_pmms')
ax[1].set_ylabel('RMSE')
ax[1].legend()
ax[0].set_title(f'Error vs k_pmm for MICE')
ax[0].set_xticks([])
ax[1].set_xticks(k_pmms)
plt.show()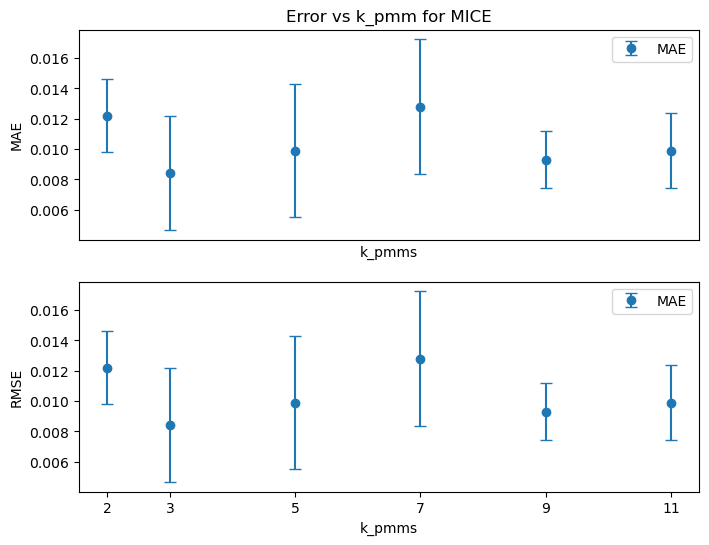
Choose k_pmm = 3 as the default for MICE.